User manual
Search
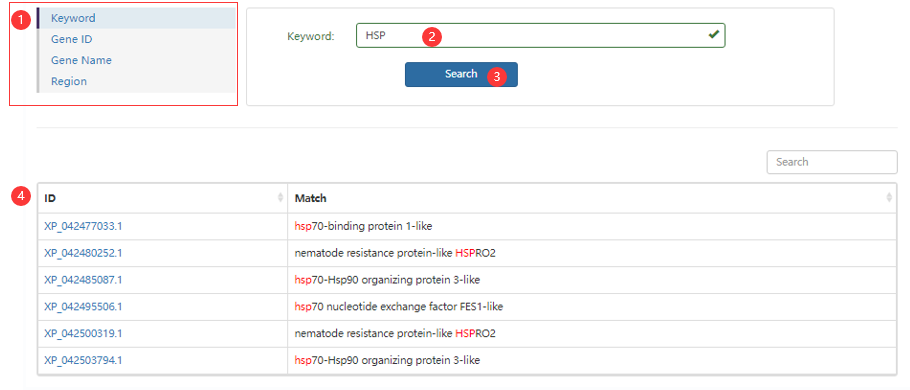
Figure1 the search page (displaying an example of Keyword type)
Step 1: choose a search type.
Step 2: enter your information.
Step 3: click search button.
Step 4: obtain a search result.
Note: you can use gene abbreviations, but it’s best to use the full gene name in keyword searches.
JBowse
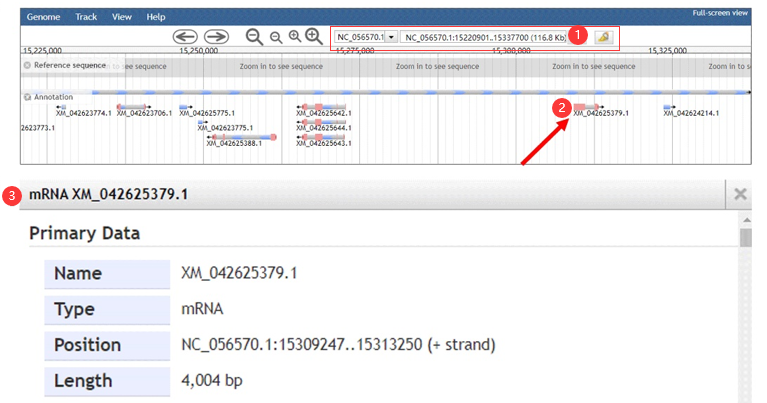
Figure 2 the JBowse page
Step 1: choose chromosome id and enter a genomic region.
Step 2: choose a target gene and click this gene.
Step 3: obtain the gene information.
Browse
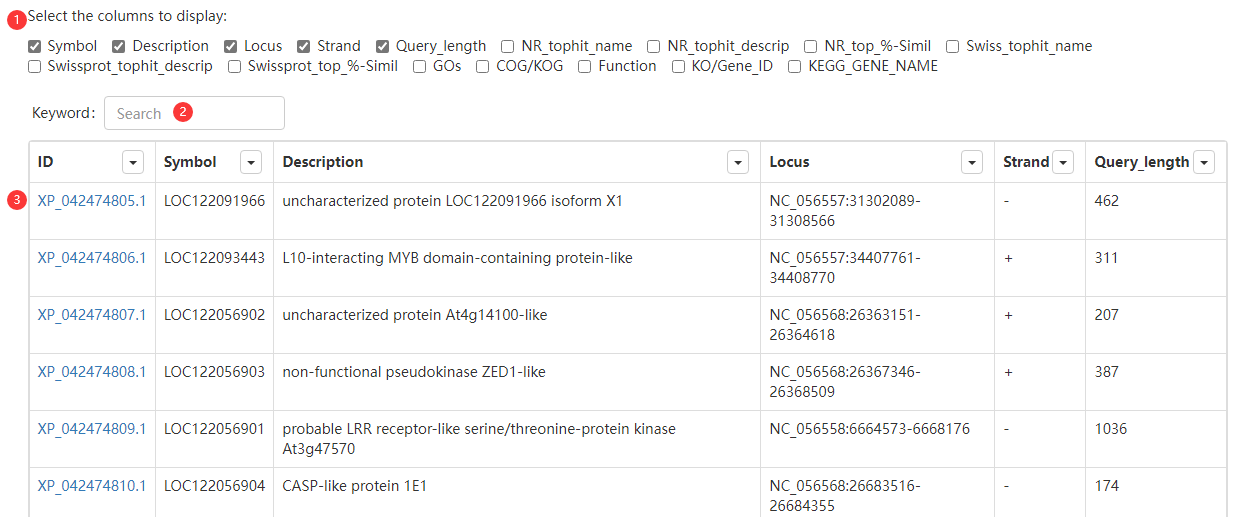
Figure 3 the Browse page
Step 1: choose information columns to display.
Step 2: enter a Keyword.
Step 3: obtain search result.
BLAST
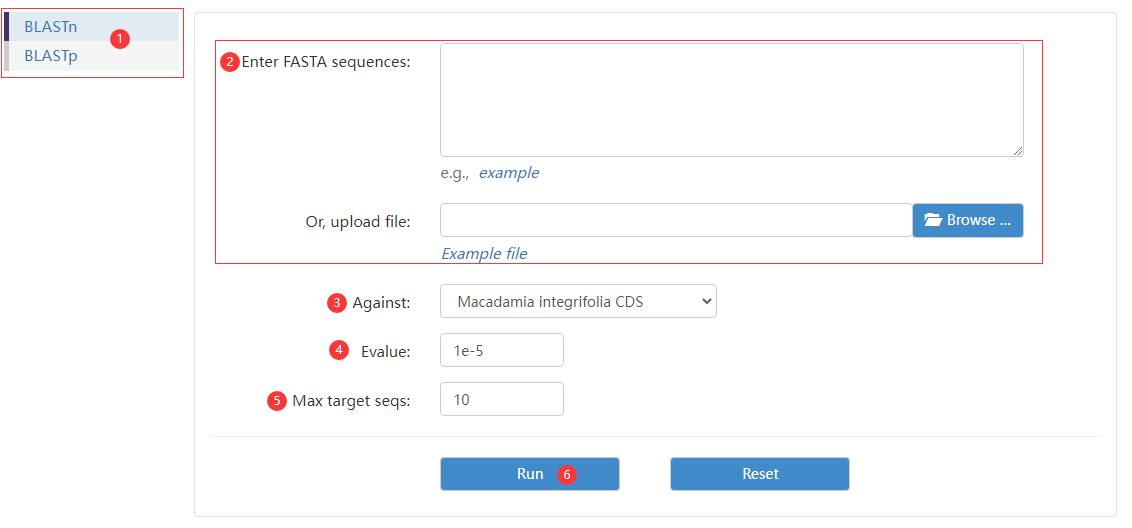
Figure 4 the BLAST page
Step 1: choose a BLAST database.
Step 2: choose a data format and enter query sequence or FASTA file.
Step 3: choose query database type.
Step 4: adjust E-value.
Step 5: choose target sequence number.
Step 6: click run button to obtain matching results.
Primer designer (Primer 3)
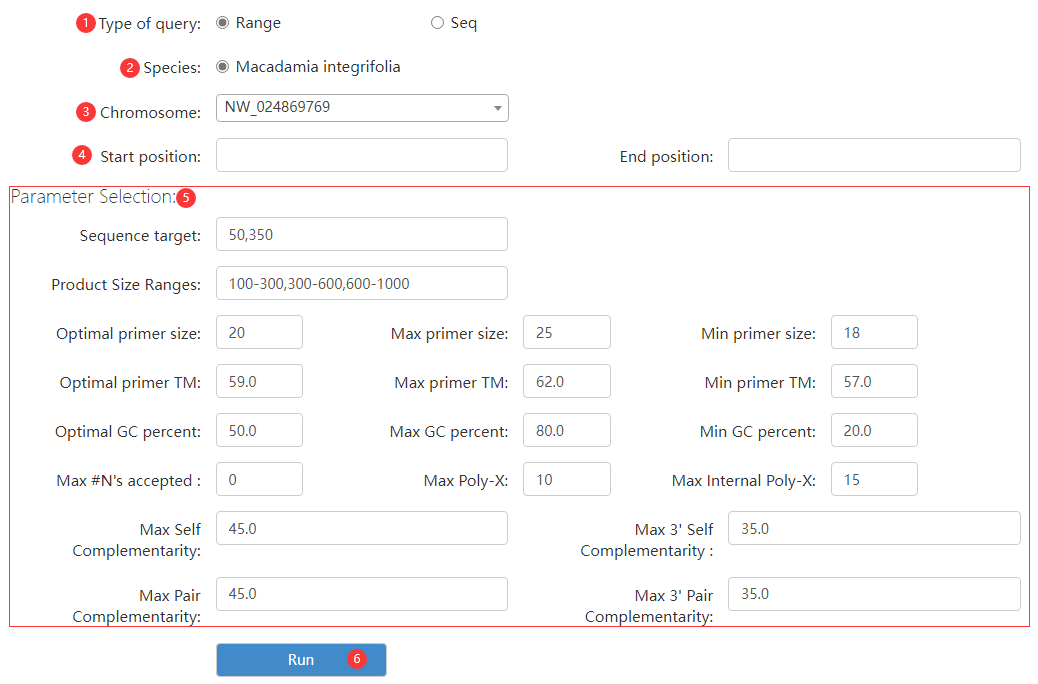
Figure 5 the Primer 3 page (displaying an example of genomic region type)
Step 1: choose a query type.
Step 2: choose a species.
Step 3: choose chromosome id.
Step 4: enter sequence position information.
Step 5: adjust primer design parameters.
Step 6: click run button to obtain candidate primers.
Sequence Fetch
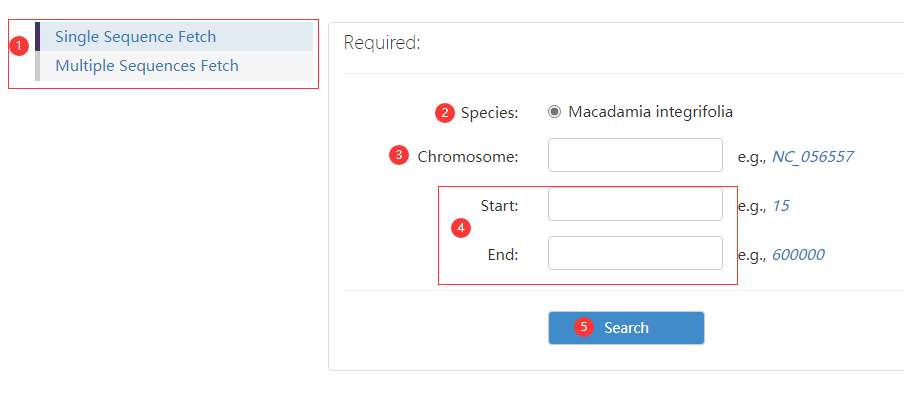
Figure 6 the Sequence Fetch page (displaying an example of single sequence fetch)
Step 1: choose a database.
Step 2: choose a species.
Step 3: choose chromosome id.
Step 4: enter sequence region information.
Step 5: click search button to obtain result.
Enrichment analysis
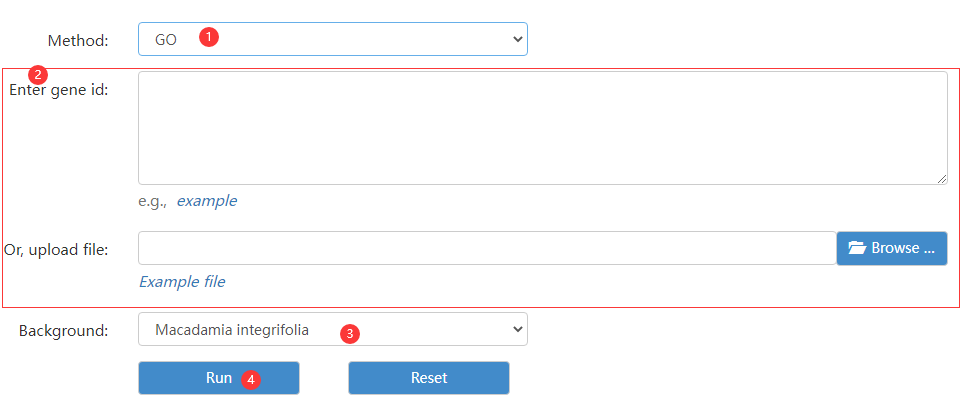
Figure 7 the Enrichment analysis page
Step 1: choose enrichment analysis database type.
Step 2: enter gene id.
Step 3: choose a background species.
Step 4: click run button to obtain enrichment result.
Multiple sequence alignment (Muscle)
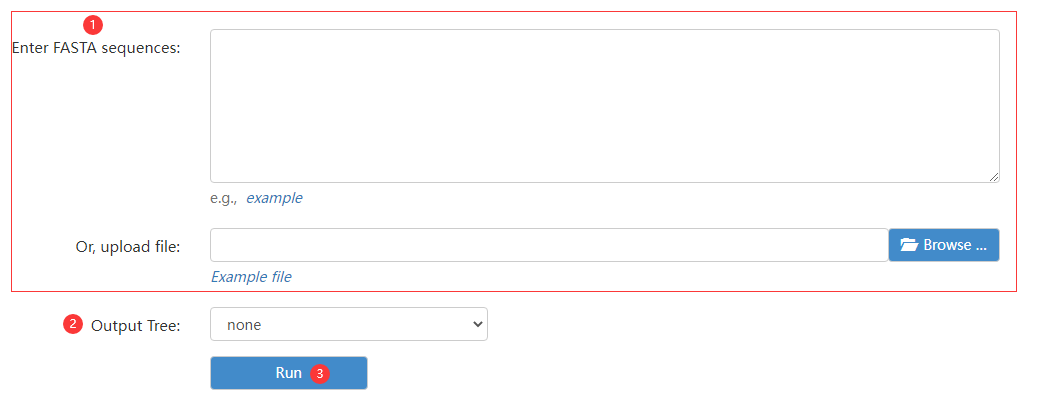
Figure 7 the Muscle page
Step 1: enter no less than two target sequence or a FASTA file.
Step 2: choose visualization.
Step 3: click run button to obtain multiple sequence alignment result.
Genome alignment (LASTZ)
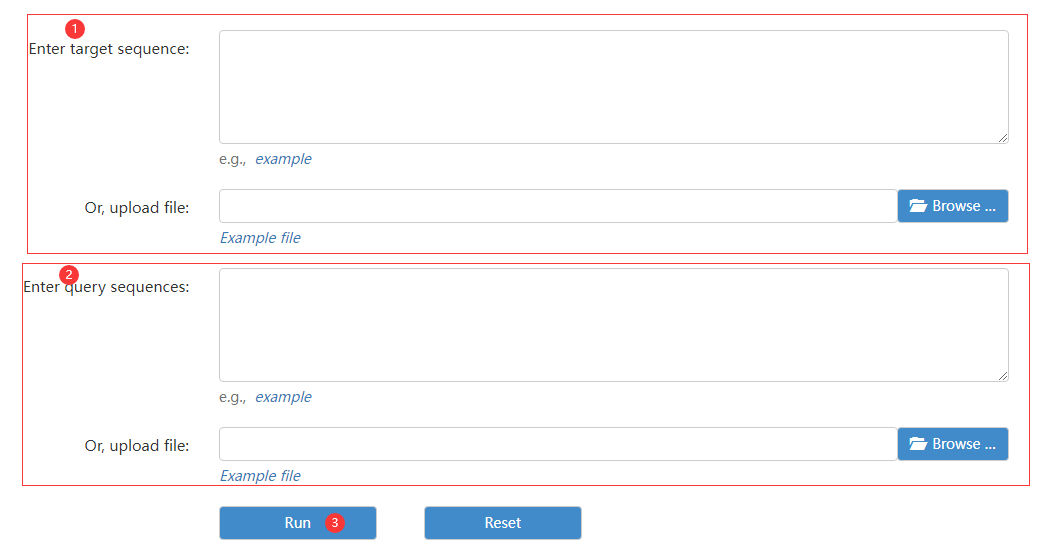
Figure 8 the LASTZ page
Step 1: enter target sequence or FATSA file.
Step 2: enter query sequence or FATSA file.
Step 3: click run to obtain genome alignment result.
Gene homology annotation (GeneWise)
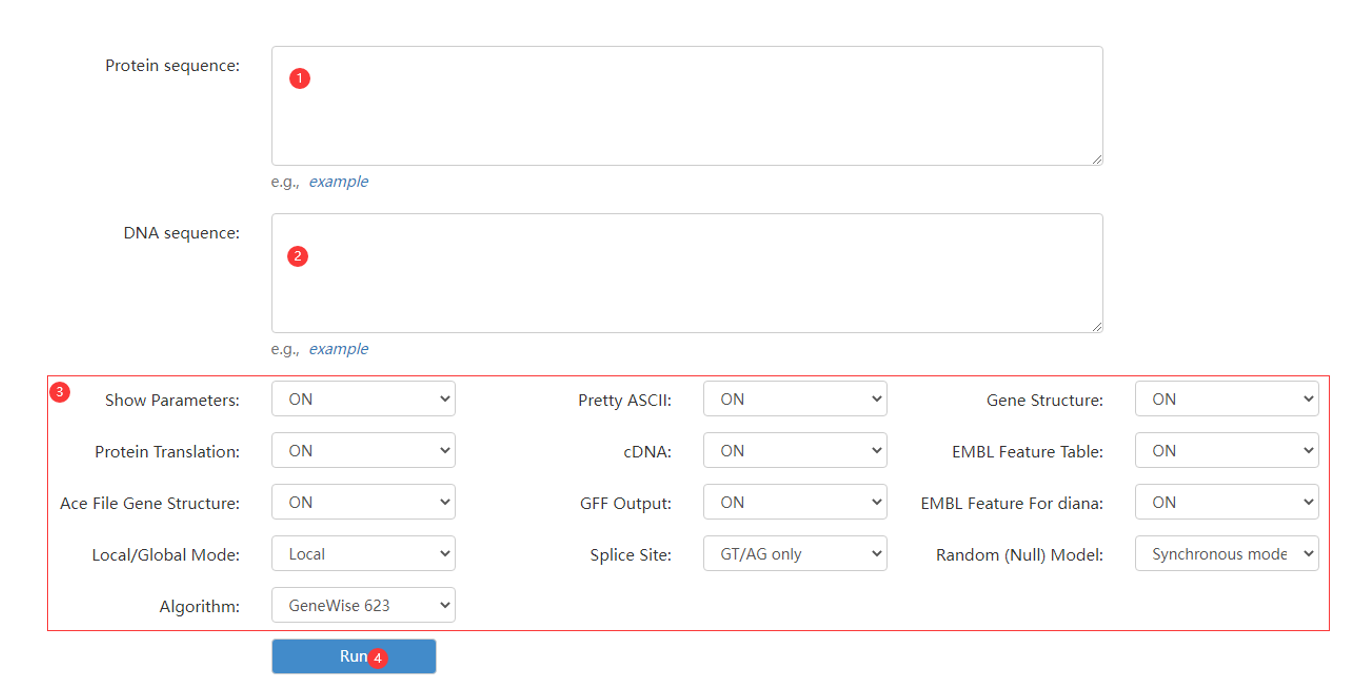
Figure 9 the GeneWise page
Step 1: enter a protein sequence.
Step 2: enter a DNA sequence.
Step 3: adjust parameters.
Step 4: click run button to obtain gene homology annotation result.
